All that salt
Friends, as you know, I've been working with my grad student lately to try and optimize our methods for extracting and sequencing DNA from larvae. The crux of the problem is that larvae are so small and yield so little DNA that you can't use a lot of the standard methods to check yourself along the way. Basically, you don't know if you've succeeded until the very, very end of the process.
We started with DNA extractions. Then we ran PCRs to copy one particular piece of DNA. It looked like we had succeeded, but then the sequences still came back wonky. It was pretty frustrating.
 |
| This is a chromatogram from Sanger sequencing of one of our PCRs. There are supposed to be nice, clean peaks that alternate for the different base pairs, but this is just...an absolute mess. |
As always, she came back almost instantly with viable, tractable solutions to try. Her main piece of advice: go easy on the salt.
You see, PCR is a chemical reaction in a tube. You add all the reagents together, expose them to the right temperatures, and magic happens. In a lot of ways, I think of PCR like baking: if you use ingredients in the wrong proportions, then things aren't going to turn out how you want. A high concentration of salts can interfere with DNA sequencing, so Hanny suggested I reduce the amount of magnesium chloride in my PCRs.
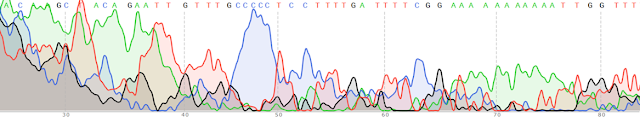
And you know what, it worked - well, at least for one of the six samples we tried. You can see the chromatogram below. All the peaks are nice and clean, and they don't overlap. The DNA sequence is given at the top. My grad student uploaded the sequence into GenBank (the go-to online database for DNA sequences) and even got a result: the tiny snail larva she had sampled was in the genus Bittium!
 |
| This is what a chromatogram is supposed to look like! |
It was incredibly satisfying to see one of our samples work. The other 5 out of 6 were still pretty wonky, but we have a few more tricks to try. With any luck, we'll have a reliable method figured out soon!
Comments
Post a Comment